1. Data input
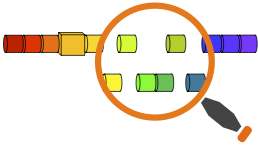
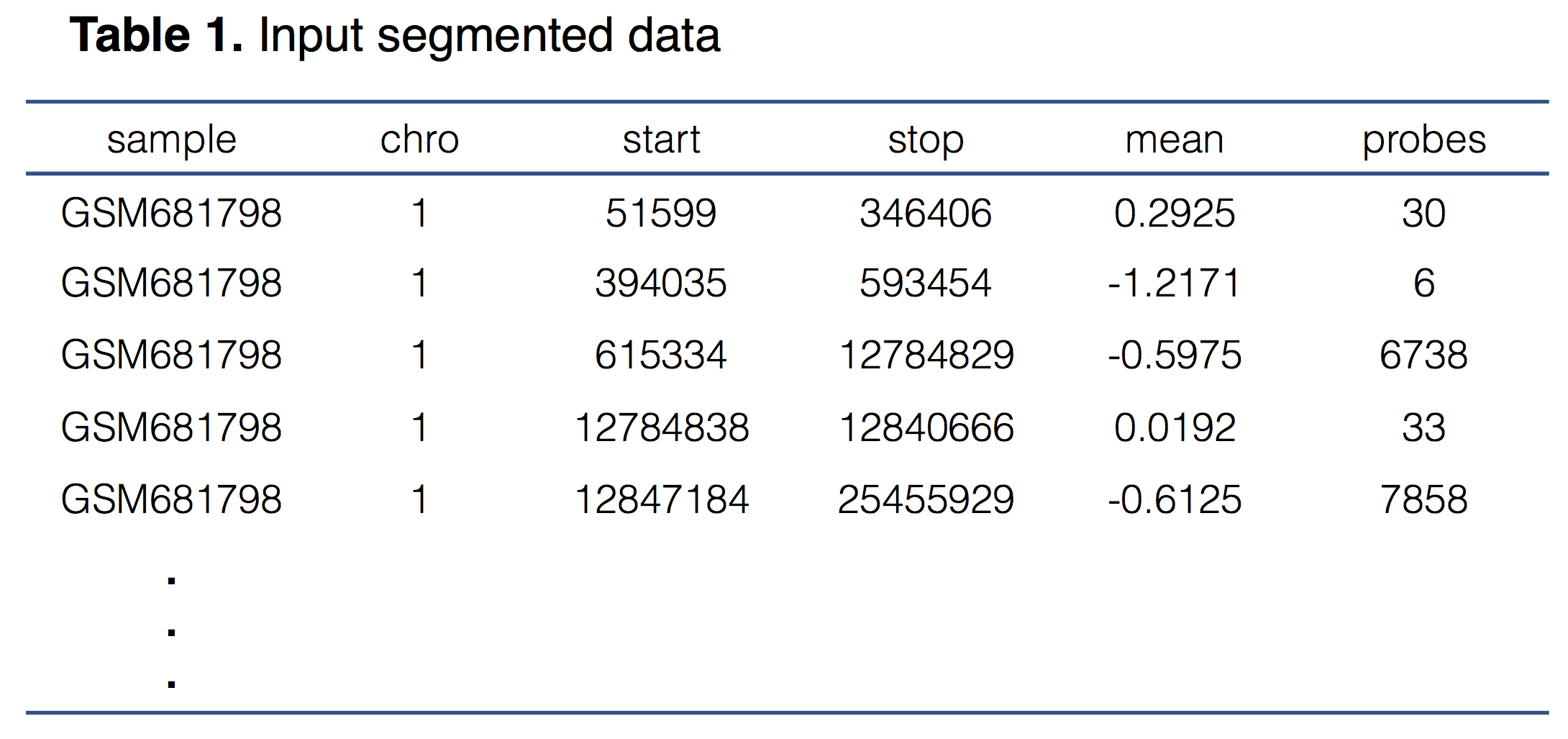
The detection of chromothripsis event is based on DNA copy number segmentation data, which can be generated by array Comparative Genomic Hybridisation (aCGH) or Single Nucleotide Polymorphism (SNP) array platforms. The minimum required data contents include the sample name, chromosome number, start and stop positions of each segment, and normalized signal intensity. The input file should be tab-delimited with an optional first line identifying the columns. Table 1 indicates an example of the input data format and content.
The uploaded data file should be plain text format with tab separator. The supported file types include: .txt, .csv, .tab, .zip. The minimum required data fields for CTLPScanner:
1. sample: The name of the data file. This name will be displayed in the figure of the result page.
2. chro: The chromosome identifier (e.g. chr1, chr6).
3. start: The starting position of the segment.
4. stop: The ending position of the segment.
5. mean: The normalized fluorescence intensity of the segment. It is usually log2 transformed.
The segment data can be “copy and paste” into the text box directly or uploaded as a flat file (Figure 1). A compressed file (.zip) can be submitted for batch processing.
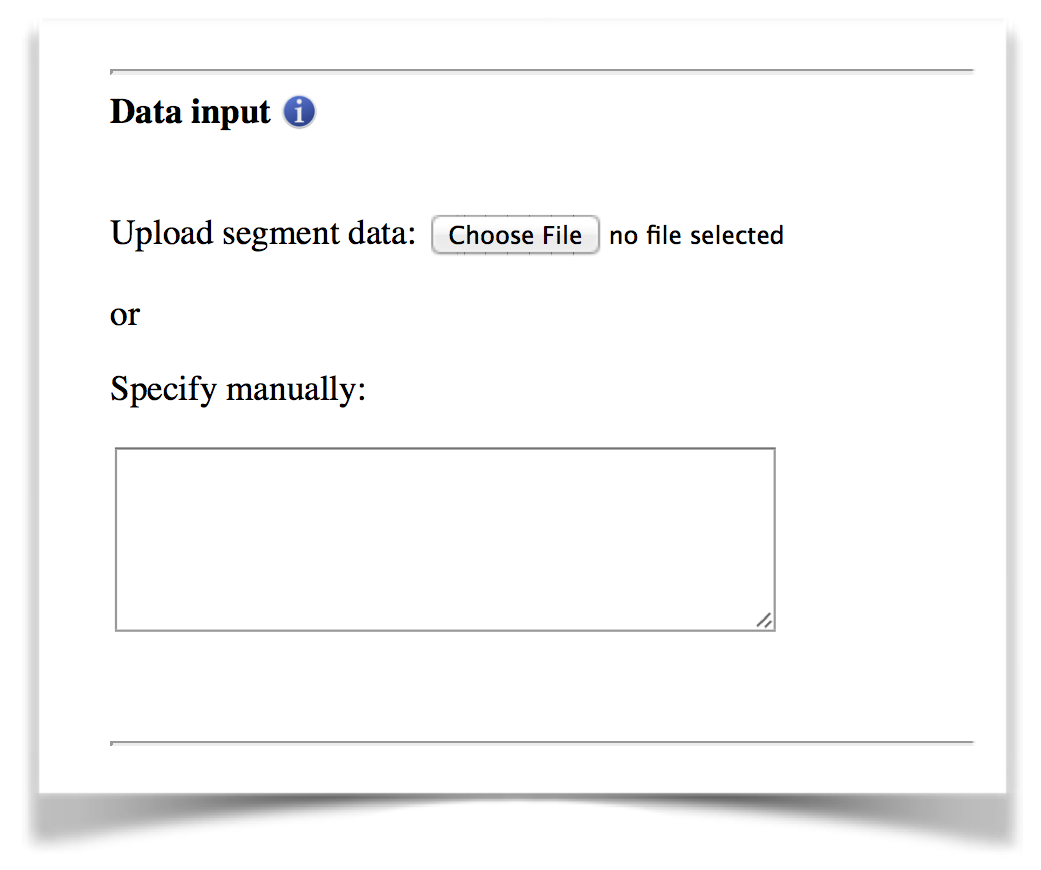
Figure 1. The data input interface.
2. Parameters & thresholds
CTLPScanner offers a set of parameters for accurate detection of chromothripsis. The web server provides optimized default values for all parameters, which may also be adjusted for customized screening (Figure 2).
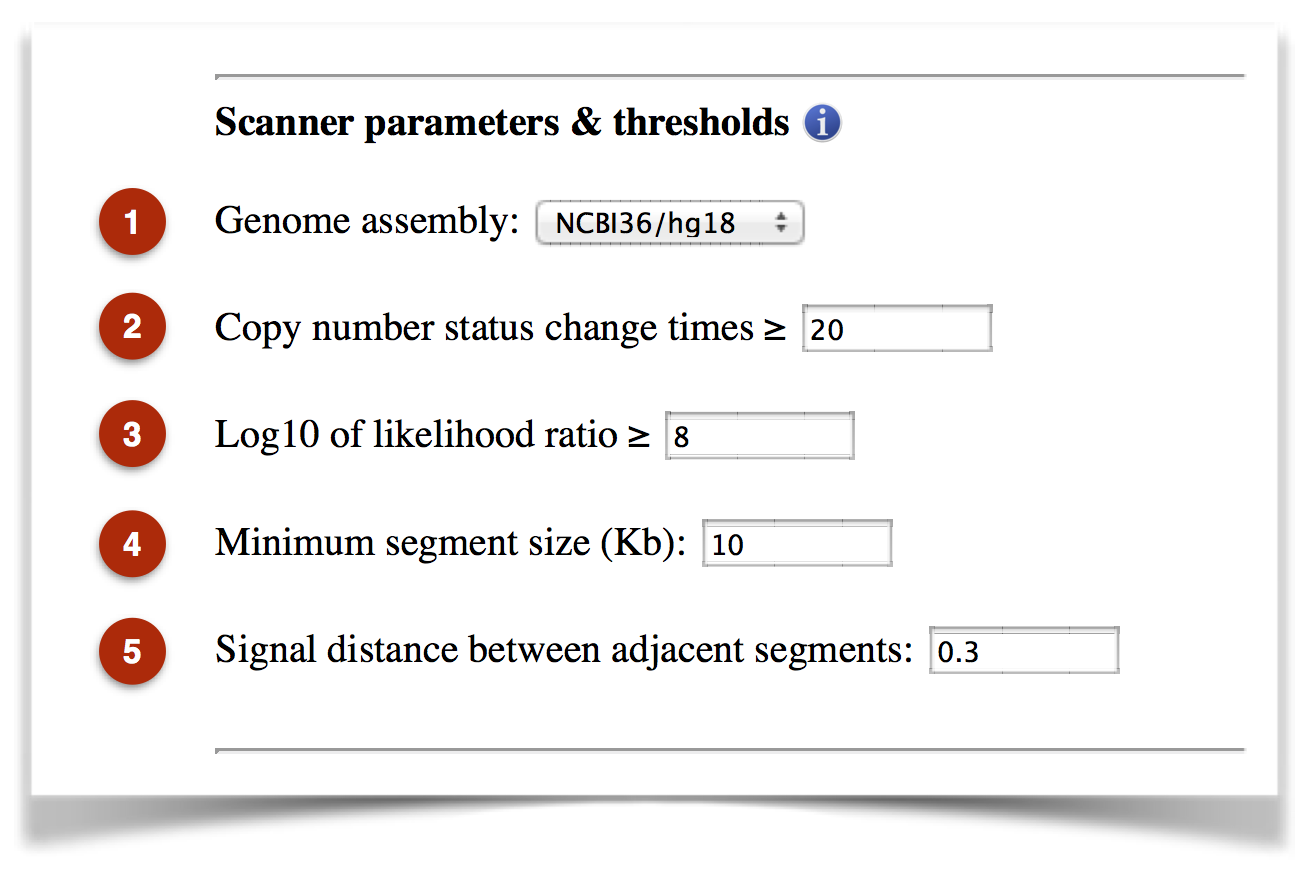
Figure 2. The parameters and thresholds for chromothripsis detection.
(1) Genome assembly: The human reference genome assembly. The available versions range from NCBI34/hg16 to NCBI38/hg38.
(2) Copy number status change times: The oscillation of copy number status change is one of the most striking features of chromothripsis. The default value is 20, which was used by the original study that reported chromothripsis phenomenon.
(3) Log10 of likelihood ratio: The clustering of chromosomal breakpoints is a key feature of chromothripsis. This parameter is used to evaluate the statistical significance of breakpoints clustering.
(4) Minimum segment size (Kb): This parameter is used to filter out the small segments that may be generated by background noise or hyper-segmentation. The default value is 10Kb, which means that segments smaller than 10 Kb will be discarded.
(5) Signal distance between adjacent segments: Due to local correlation effects between probes or the existence of background noise, the segmentation profiles occasionally exhibit subtle striation patterns. This pattern is constituted with a large number of small segments, which is unlikely to be a biological phenomenon. To reduce artificial copy number status change, the distance of signal intensity between adjacent segments was introduced. If the distance of two adjacent segments differed by less than this threshold, the copy number status change was not considered.
The customized visualization parameters are provided by CTLPScanner (Figure 3).

Figure 3. The visualization options.
(1) The thresholds for calling genomic gains and losses: These values are used to “call” genomic gains and losses. The default values are 0.15 and -0.15, which are commonly used across multiple array platforms.
(2) Color for genomic gains or losses: The colors used to indicate copy number gains and losses.
(3) Show detected chromothripsis-like region only: If the check box is checked, only the pulverized chromosome will be displayed.
3. Output
Parameters & Thresholds: The parameters and thresholds defined by users to perform chromothripsis detection. Users can reuse these parameters for chromothripsis detection or re-analyze the same sample with changed thresholds (Figure 4).
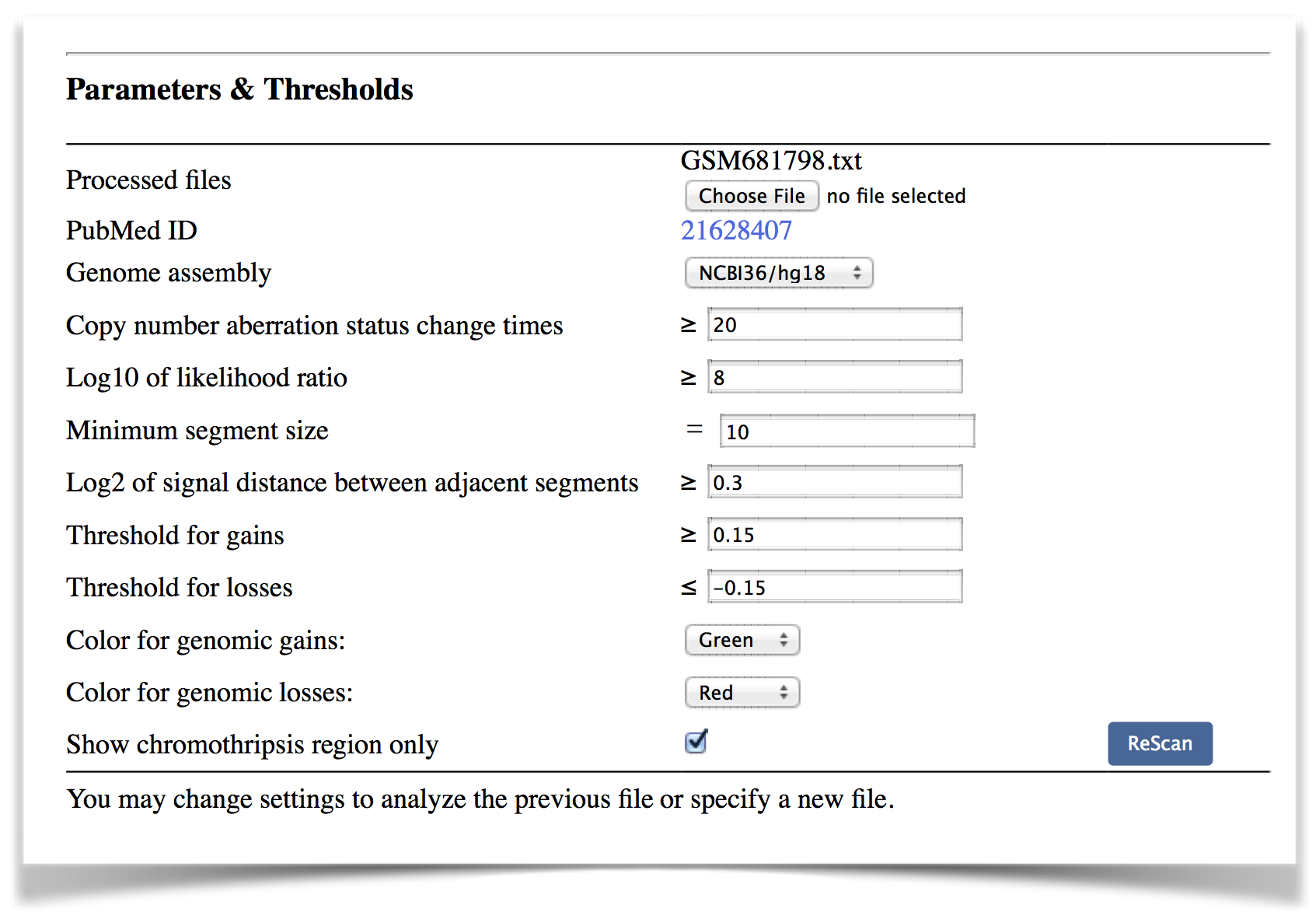
Figure 4. The parameters and thresholds used are shown on the result page.
Chromothripsis Region: In identified chromothripsis region, the two main features of chromothripsis (CNA status change times & Likelihood ratio) are shown (Figure 5). The result file can be downloaded.
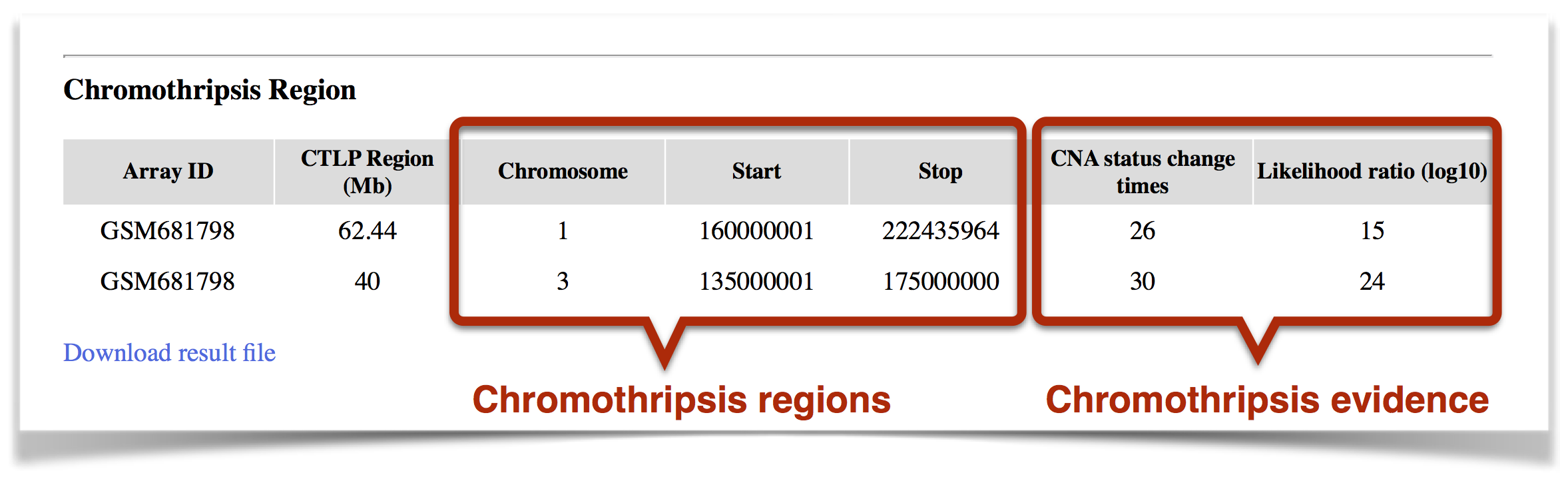
Figure 5. The detected chromothripsis region.
Chromothripsis & Copy Number Aberrations Plots:
If a chromothripsis event is detected, a general view of copy number aberration profile of the sample will be shown (Figure 6). In the plot for each chromosome, the chromothripsis region will be highlighted with a light orange rectangle (Figure 7).
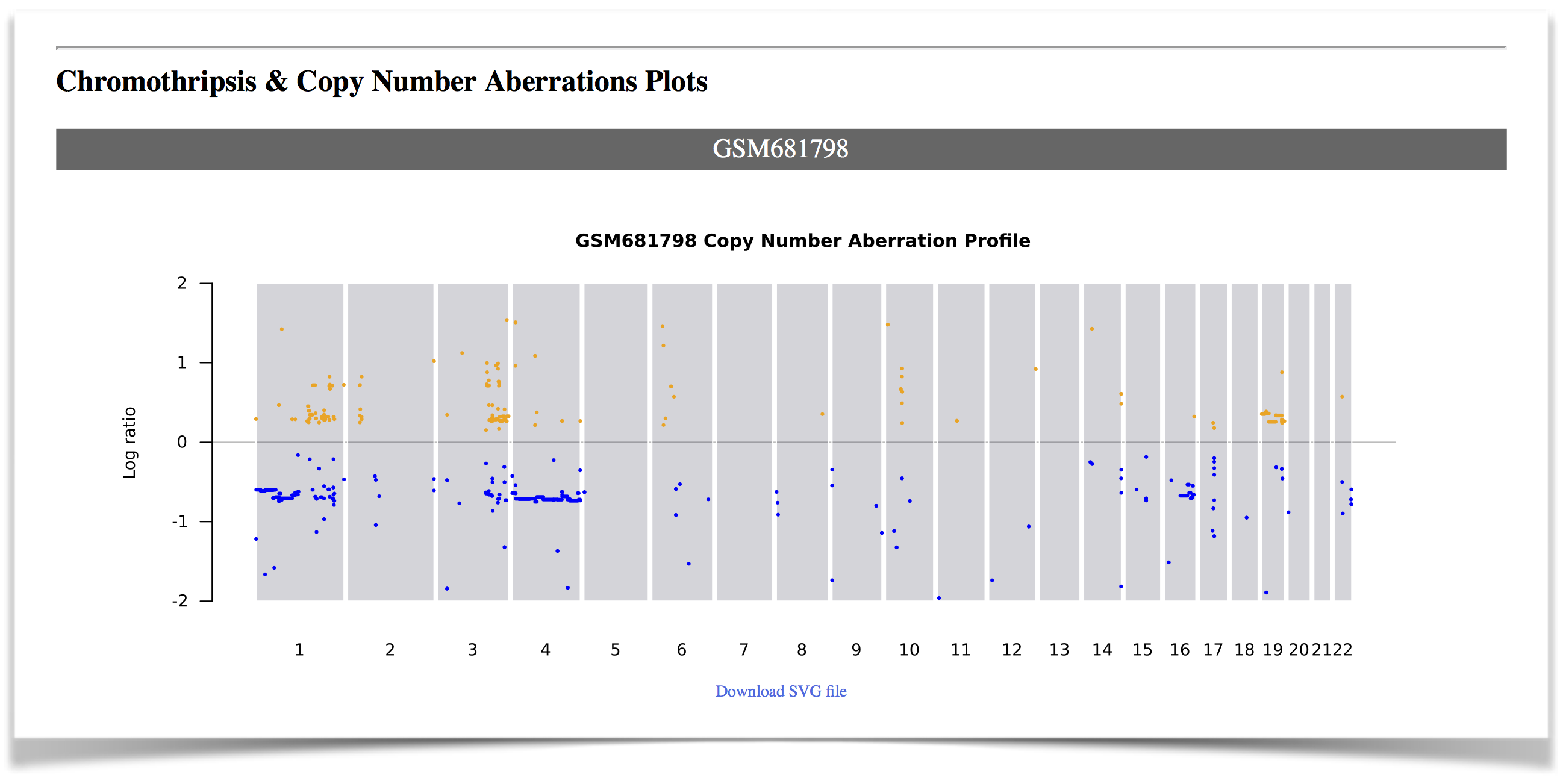
Figure 6. The general view of copy number aberration profile .
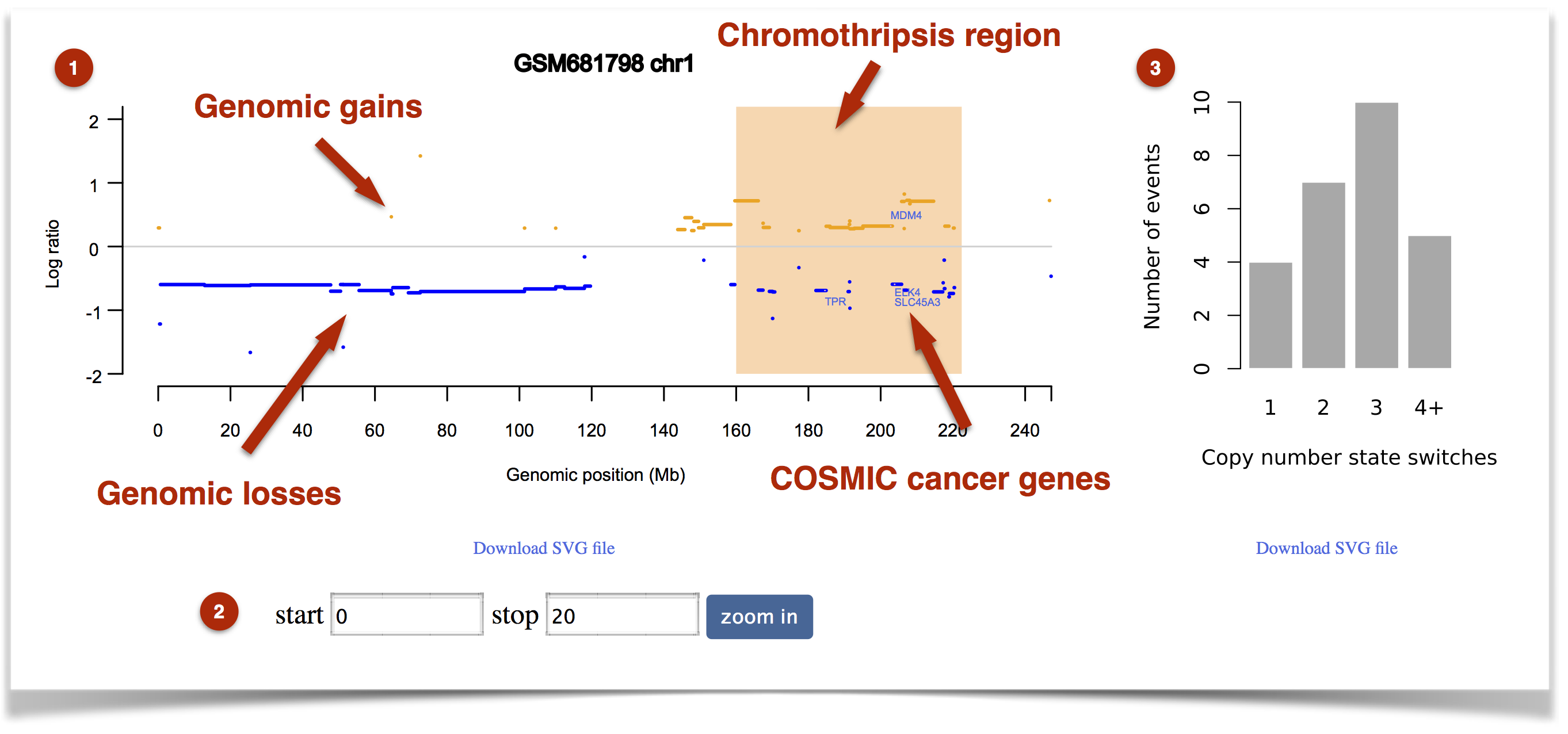
Figure 7. An example of result visualization.
(1) For each chromosome, the “called” genomic gains and losses are shown according to user-defined colors. The X and Y axes represent genomic positions and log ratio of segments, respectively. The orange rectangle represents identified chromothripsis regions.
(2) Users can zoom in specific genomic regions by inputting the start and stop locations.
(3) The histogram of copy number segment switches. It facilitates the identification of chromothripsis event arising in conjunction with telomere crises. The patterns and distribution of copy number state switches indicate the emergence and temporal order of the major genomic rearrangement events.
Cancer Genes: COSMIC (Catalogue Of Somatic Mutations In Cancer) cancer genes that are involved in the detected chromothripsis regions (Figure 8). These genes are affected by genomic gain or loss, which are represented by user-defined colors. The "segment size" indicates the size of copy number aberrations that the gene involved in. In general, the relatively small CNAs, which known as "focus changes", are in favour of identifying "driver" cancer genes.
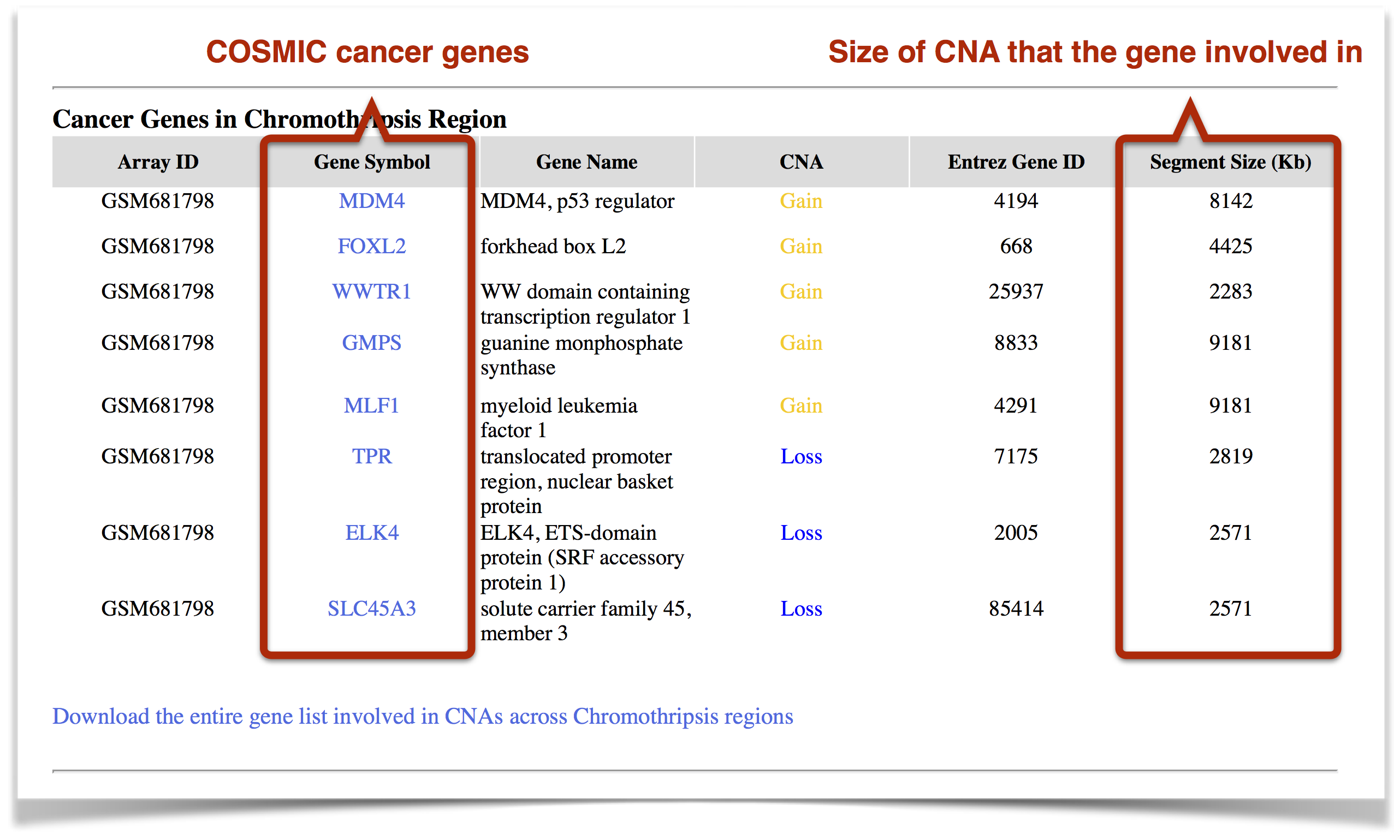
Figure 8. The cancer genes that are involved in the detected chromothripsis regions.
Scatter Plot: CTLPScanner creates a scatter plot to compare the two decision parameters, i.e. copy number status change times and likelihood ratio (Figure 9). Each point in the plot represents a chromosome, and the candidates falling in the upper right area are inferred chromothripsis cases. The closer the point is to the top-right corner, the more accurately the algorithm can distinguish chromothripsis from non-chromothripsis cases.
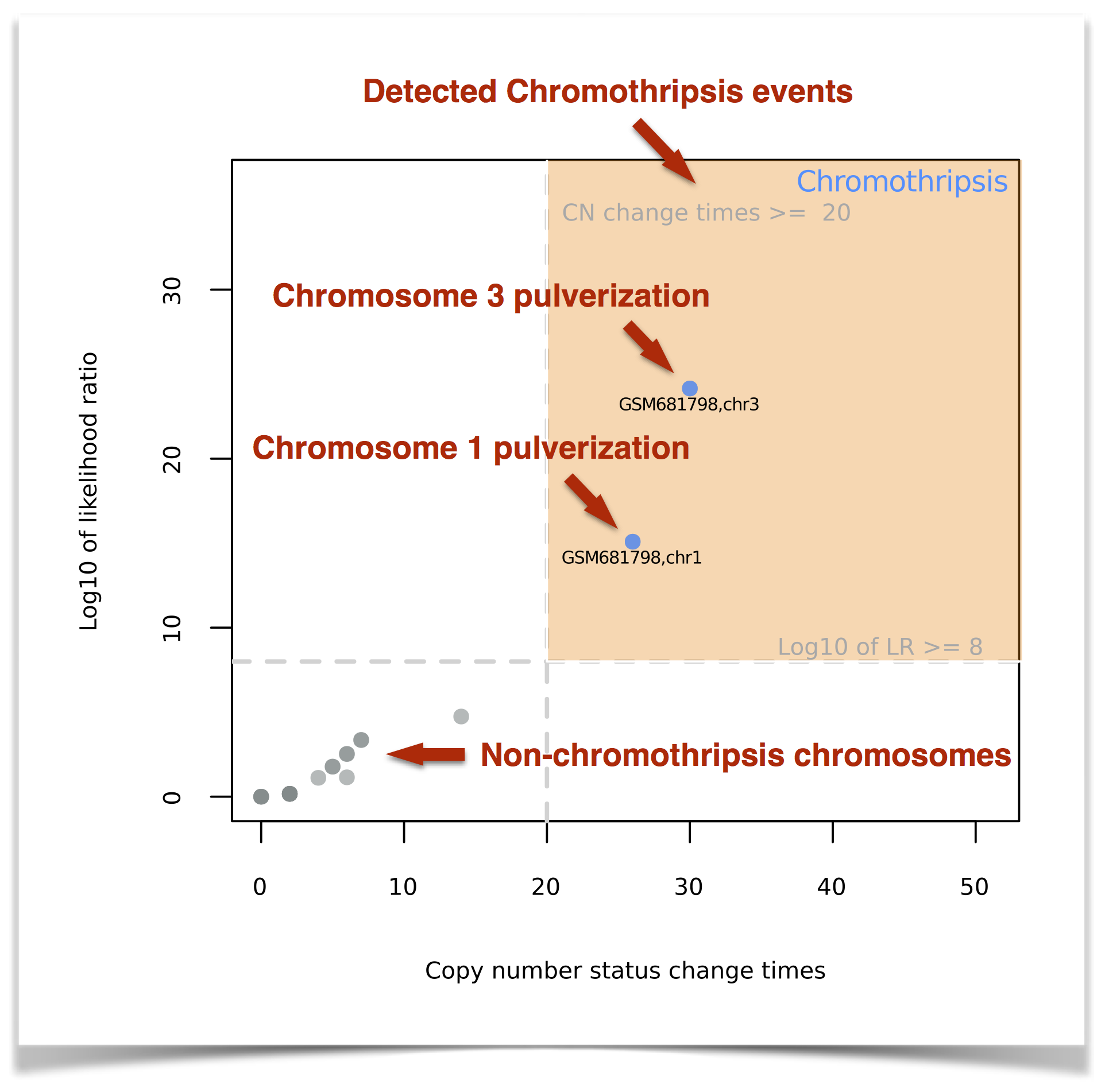
Figure 9. The scatter plot of detected chromothripsis events.
|