| |
Chromothripsis |
“Chromothripsis” was initially found in cancer cells, and was defined by the occurrence of tens of hundreds of clustered genomic rearrangements, supposedly having arisen in a single catastrophic event. In this event, contiguous chromosomal regions are fragmented into many pieces via presently unknown mechanisms. The cell’s DNA repair machinery randomly fuses these segments together to rescue the genome. These somatically acquired genomic rearrangements may result in complex patterns of regional copy number changes. They have the potential to interrupt or activate multiple genes, and are consequently implicated in cancer development. It has been proposed that this “shattering” and aberrant repair of a multitude of DNA fragments may provide an alternative oncogenetic route, in contrast to the step-by-step paradigm of cancer development. Figure 1 is a schema of chromothripsis that occurs in a single catastrophic chromosome shattering event. Recent studies have reported this phenomenon in numerous cancer types. Besides human cancers, recent research have also reported chromothripsis events in germline and non-human genomes. The discovery of chromothripsis reveals a possible new paradigm in cancer development and stimulates great interest in the study of this phenomenon. |
|
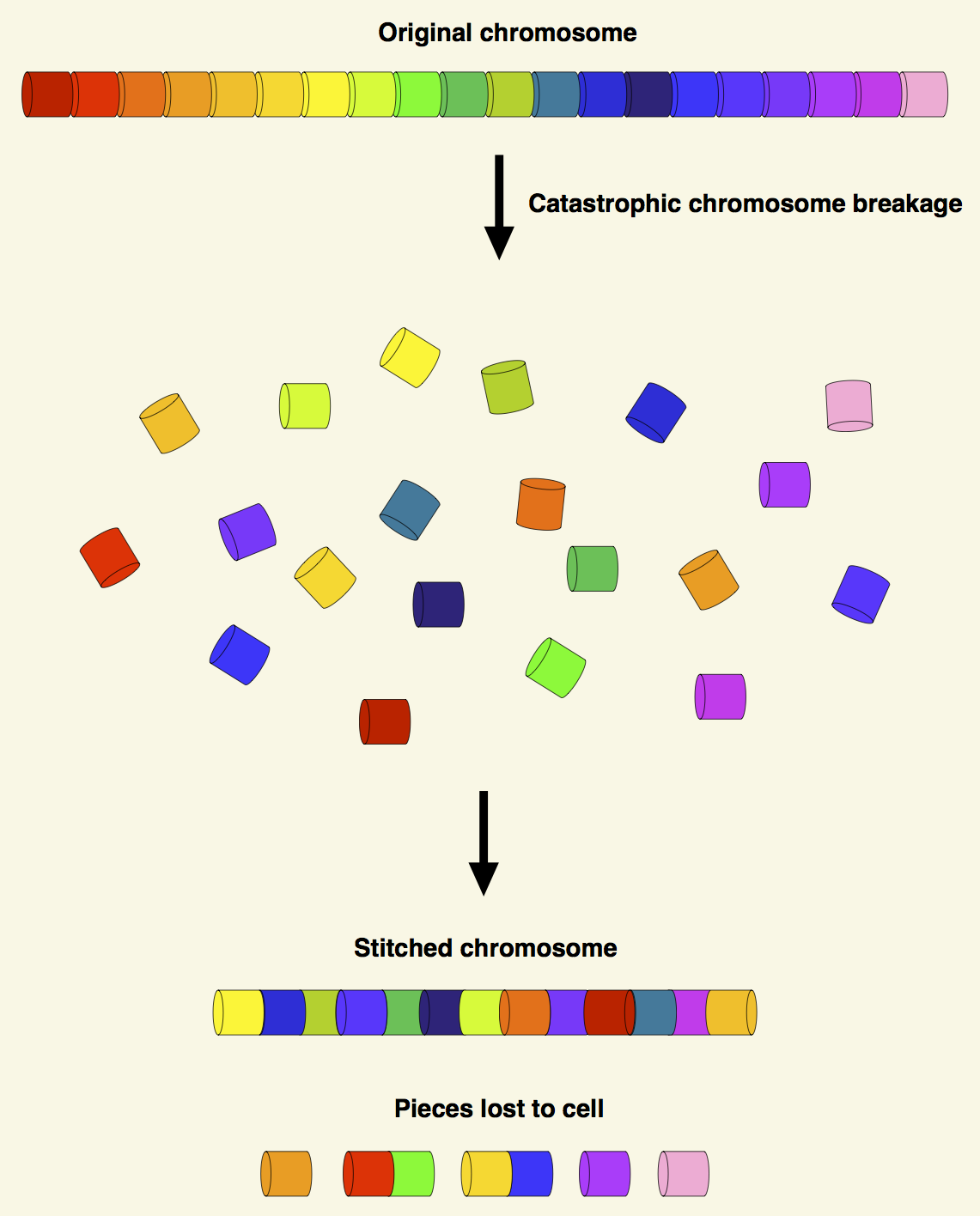
Figure 1. Schema of the process of chromothripsis in a single chromosome. The chromosome is shattered into many pieces. Then these fragments, or some of the fragments, are pieced together to generate a derivative chromosome. The other segments lost to cell. Chromothripsis can involve several chromosomes. |
Chromothripsis has been characterized as a type of focally clustered genomic aberrations generated in a one time cellular event and being limited to a defined set of copy number states. Other operational definitions have also been employed based on clustered aberrations. Chromothripsis patterns can be detected by genomic arrays. A lot of studies have confirmed the occurrence of chromothripsis in genomic array data sets. |
| |
|
 |
|
 |
| |
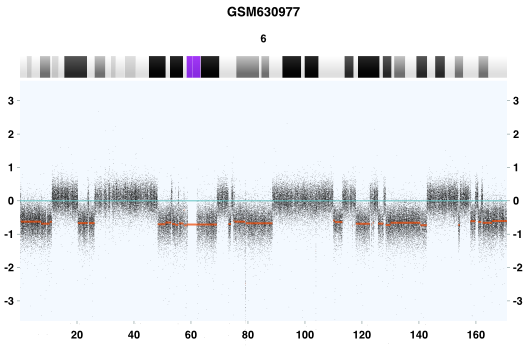
Figure 2. Chromothripsis event in the whole chromosome 6. GSM630977, Acute Myeloid Leukemia (AML), Affymetrix SNP6 platform. The x-axis indicates genomic location in Mb, and the y-axis is the log2 value of probe signal intensity. Green and red lines represent genomic gains and losses respectively.
|
|
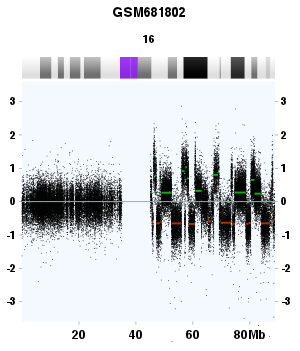
Figure 3. Chromothripsis event in the q arm of chromosome 16. GSM681802, Multiple myeloma, Affymetrix SNP6 platform. The q arm of chromosome 16 is "smashed" while the p arm is normal. The x and y axes are the same as Figure 2.
|
Chromothripsis Identification |
The accurate identification of chromothripsis regions is the basis for further investigation of the underlying mechanism. CTLPScanner aims to accurately identify chromothripsis-like events and regions in cancer genome. According to previous studies, segmental copy number status changes and significant breakpoint clustering are two relevant features of chromothripsis. For an automatic identification of CTLP, a scan-statistic likelihood ratio based on the Poisson model was employed. It is commonly used to detect clusters of events in time and/or space and to determine their statistical significance. For each chromosome, the algorithm uses a series of sliding windows to identify the genomic region with the highest likelihood ratio as the CTLP candidate. This algorithm can automatically adjust the size of “scan window” to find the reasonable region of chromothripsis. |
References |
|
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011 Jan 7;144(1):27-40.
Cai H, Kumar N, Bagheri HC, von Mering C, Robinson MD, Baudis M. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genomics. 2014 Jan 29;15:82. doi: 10.1186/1471-2164-15-82.
Cai H, Kumar N, Baudis M. arrayMap: a reference resource for genomic copy number imbalances in human malignancies. PLoS One. 2012;7(5):e36944. doi: 10.1371/journal.pone.0036944. Epub 2012 May 18.
|
|