| PubMed ID |
|
25079552 |
Publish Date |
|
2014 Jul |
| Journal |
|
Nature |
Species |
|
Homo sapiens |
| Disease Type |
|
Non Small-cell lung cancer |
Technology |
|
Next Generation Sequencing |
| Case Number |
|
6 |
Raw Data |
|
|
| Title |
|
Comprehensive molecular profiling of lung adenocarcinoma |
| Authors |
|
Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L, Ding L, Getz G, Hammerman PS, Hayes DN, Hernandez B, Herman JG, Heymach JV, Jurisica I, Kucherlapati R, Kwiatkowski D, Ladanyi M, Robertson G, Schultz N, Shen R, Sinha R, Sougnez C, Tsao MS, Travis WD, Weinstein JN, Wigle DA, Wilkerson MD, Chu A, Cherniack AD, Hadjipanayis A, Rosenberg M, Weisenberger DJ, Laird PW, Radenbaugh A, Ma S, Stuart JM, Averett Byers L, Baylin SB, Govindan R, Meyerson M, Rosenberg M, Gabriel SB, Cibulskis K, Sougnez C, Kim J, Stewart C, Lichtenstein L, Lander ES, Lawrence MS, Getz, Kandoth C, Fulton R, Fulton LL, McLellan MD, Wilson RK, Ye K, Fronick CC, Maher CA, Miller CA, Wendl MC, Cabanski C, Ding L, Mardis E, Govindan R, Creighton CJ, Wheeler D, Balasundaram M, Butterfield YS, Carlsen R, Chu A, Chuah E, Dhalla N, Guin R, Hirst C, Lee D, Li HI, Mayo M, Moore RA, Mungall AJ, Schein JE, Sipahimalani P, Tam A, Varhol R, Robertson A, Wye N, Thiessen N, Holt RA, Jones SJ, Marra MA, Campbell JD, Brooks AN, Chmielecki J, Imielinski M, Onofrio RC, Hodis E, Zack T, Sougnez C, Helman E, Sekhar Pedamallu C, Mesirov J, Cherniack AD, Saksena G, Schumacher SE, Carter SL, Hernandez B, Garraway L, Beroukhim R, Gabriel SB, Getz G, Meyerson M, Hadjipanayis A, Lee S, Mahadeshwar HS, Pantazi A, Protopopov A, Ren X, Seth S, Song X, Tang J, Yang L, Zhang J, Chen PC, Parfenov M, Wei Xu A, Santoso N, Chin L, Park PJ, Kucherlapati R, Hoadley KA, Auman JT, Meng S, Shi Y, Buda E, Waring S, Veluvolu U, Tan D, Mieczkowski PA, Jones CD, Simons JV, Soloway MG, Bodenheimer T, Jefferys SR, Roach J, Hoyle AP, Wu J, Balu S, Singh D, Prins JF, Marron JS, Parker JS, Hayes DN, Perou CM, Liu J, Cope L, Danilova L, Weisenberger DJ, Maglinte DT, Lai PH, Bootwalla MS, Van Den Berg DJ, Triche T Jr, Baylin SB, Laird PW, Rosenberg M, Chin L, Zhang J, Cho J, DiCara D, Heiman D, Lin P, Mallard W, Voet D, Zhang H, Zou L, Noble MS, Lawrence MS, Saksena G, Gehlenborg N, Thorvaldsdottir H, Mesirov J, Nazaire MD, Robinson J, Getz G, Lee W, Aksoy BA, Ciriello G, Taylor BS, Dresdner G, Gao J, Gross B, Seshan VE, Ladanyi M, Reva B, Sinha R, Sumer SO, Weinhold N, Schultz N, Shen R, Sander C, Ng S, Ma S, Zhu J, Radenbaugh A, Stuart JM, Benz CC, Yau C, Haussler D, Spellman PT, Wilkerson MD, Parker JS, Hoadley KA, Kimes PK, Hayes DN, Perou CM, Broom BM, Wang J, Lu Y, Kwok Shing Ng P, Diao L, Averett Byers L, Liu W, Heymach JV, Amos CI, Weinstein JN, Akbani R, Mills GB, Curley E, Paulauskis J, Lau K, Morris S, Shelton T, Mallery D, Gardner J, Penny R, Saller C, Tarvin K, Richards WG, Cerfolio R, Bryant A, Raymond DP, Pennell NA, Farver C, Czerwinski C, Huelsenbeck-Dill L, Iacocca M, Petrelli N, Rabeno B, Brown J, Bauer T, Dolzhanskiy O, Potapova O, Rotin D, Voronina O, Nemirovich-Danchenko E, Fedosenko KV, Gal A, Behera M, Ramalingam SS, Sica G, Flieder D, Boyd J, Weaver J, Kohl B, Huy Quoc Thinh D, Sandusky G, Juhl H, Duhig E, Illei P, Gabrielson E, Shin J, Lee B, Rodgers K, Trusty D, Brock MV, Williamson C, Burks E, Rieger-Christ K, Holway A, Sullivan T, Wigle DA, Asiedu MK, Kosari F, Travis WD, Rekhtman N, Zakowski M, Rusch VW, Zippile P, Suh J, Pass H, Goparaju C, Owusu-Sarpong Y, Bartlett JM, Kodeeswaran S, Parfitt J, Sekhon H, Albert M, Eckman J, Myers JB, Cheney R, Morrison C, Gaudioso C, Borgia JA, Bonomi P, Pool M, Liptay MJ, Moiseenko F, Zaytseva I, Dienemann H, Meister M, Schnabel PA, Muley TR, Peifer M, Gomez-Fernandez C, Herbert L, Egea S, Huang M, Thorne LB, Boice L, Hill Salazar A, Funkhouser WK, Rathmell WK, Dhir R, Yousem SA, Dacic S, Schneider F, Siegfried JM, Hajek R, Watson MA, McDonald S, Meyers B, Clarke B, Yang IA, Fong KM, Hunter L, Windsor M, Bowman RV, Peters S, Letovanec I, Khan KZ, Jensen MA, Snyder EE, Srinivasan D, Kahn AB, Baboud J, Pot DA, Mills Shaw KR, Sheth M, Davidsen T, Demchok JA, Yang L, Wang Z, Tarnuzzer R, Zenklusen JC, Ozenberger BA, Sofia HJ, Travis WD, Cheney R, Clarke B, Dacic S, Duhig E, Funkhouser WK, Illei P, Farver C, Rekhtman N, Sica G, Suh J, Tsao MS, Travis WD, Cheney R, Clarke B, Dacic S, Duhig E, Funkhouser WK, Illei P, Farver C, Rekhtman N, Sica G, Suh J, Tsao MS |
| Affiliation |
|
University of CaliforniaSan Francisco, San Francisco,California94158,USA |
| Chromothripsis Definition |
|
Close-by breakpoints: >=2
Copy number states: 2-3
Fragments random joining: Yes |
| Download |
|
Study Data File |
| |
| Chromothripsis Cases |
| TCGA-05-5715 |
| |
|
|
| |
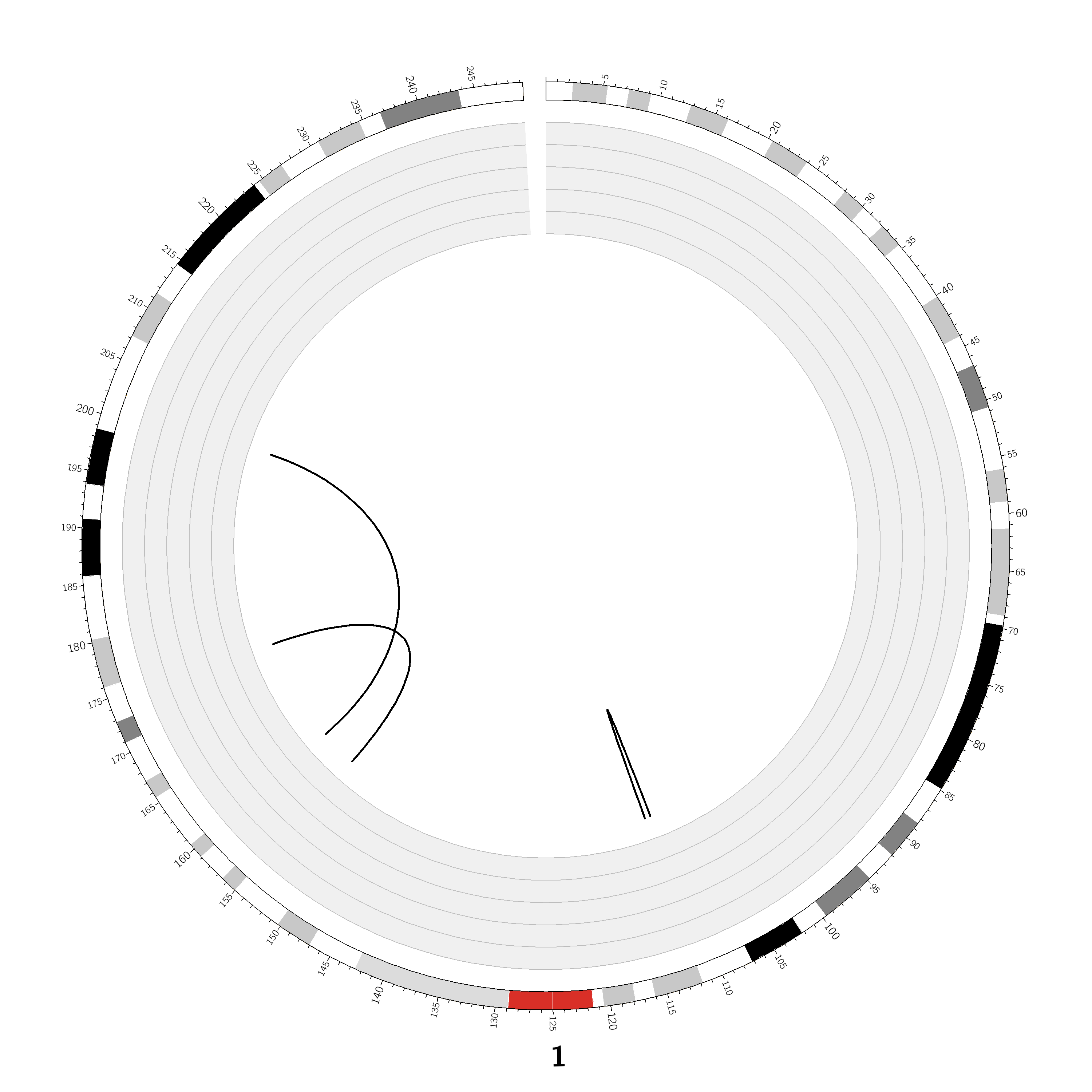
Case ID: |
TCGA-05-5715 |
| |
Chromosome: | 1 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| |
Affected Gene: |
Gene List |
| |
Download: |
Links File |
| |  | | TCGA-50-5932 |
| |
|
|
| |
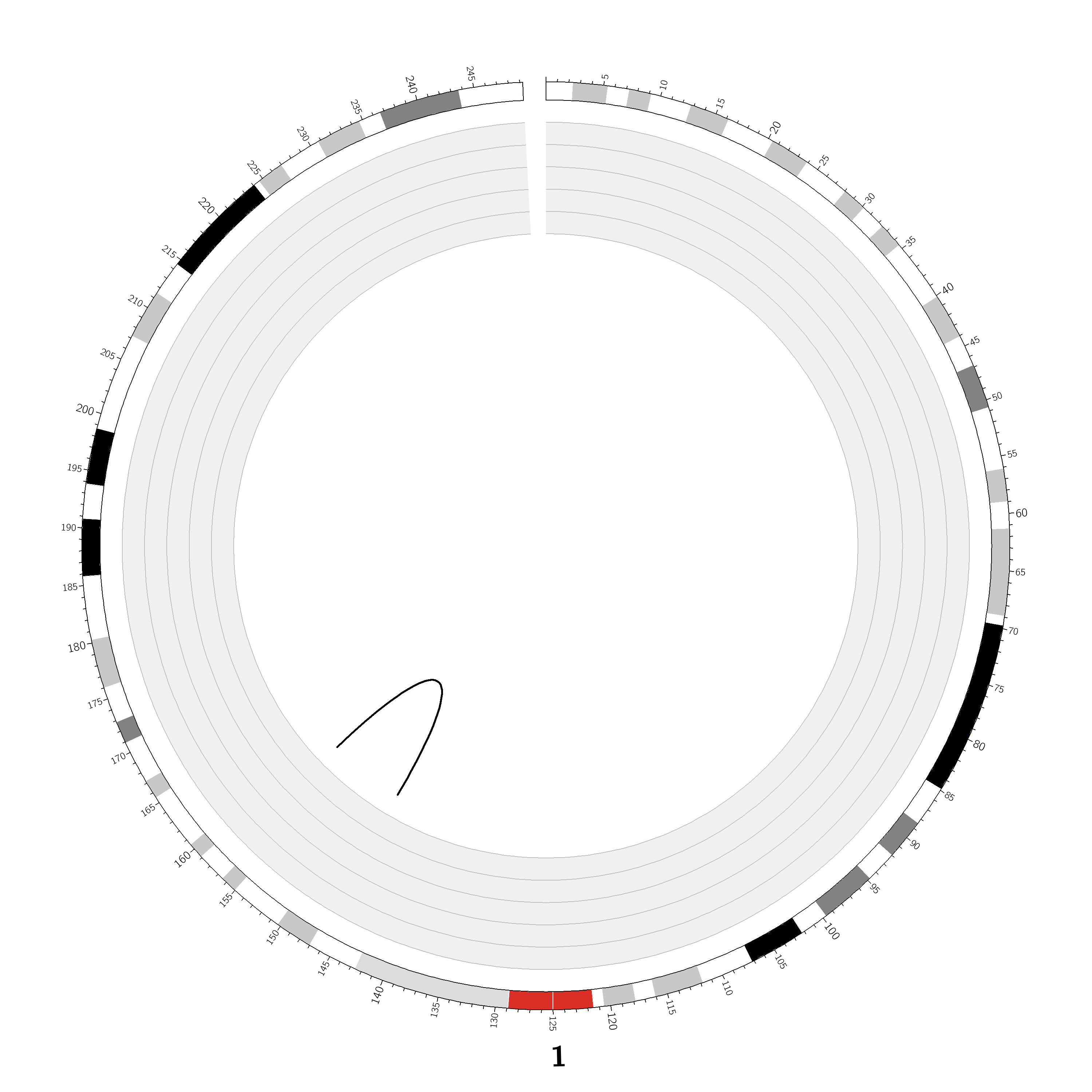
Case ID: |
TCGA-50-5932 |
| |
Chromosome: | 1 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| |
Affected Gene: |
Gene List |
| |
Download: |
Links File |
| |  | | TCGA-67-3771 |
| |
|
|
| |
Case ID: |
TCGA-67-3771 |
| |
Chromosome: | 3 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| | | TCGA-49-4488 |
| |
|
|
| |
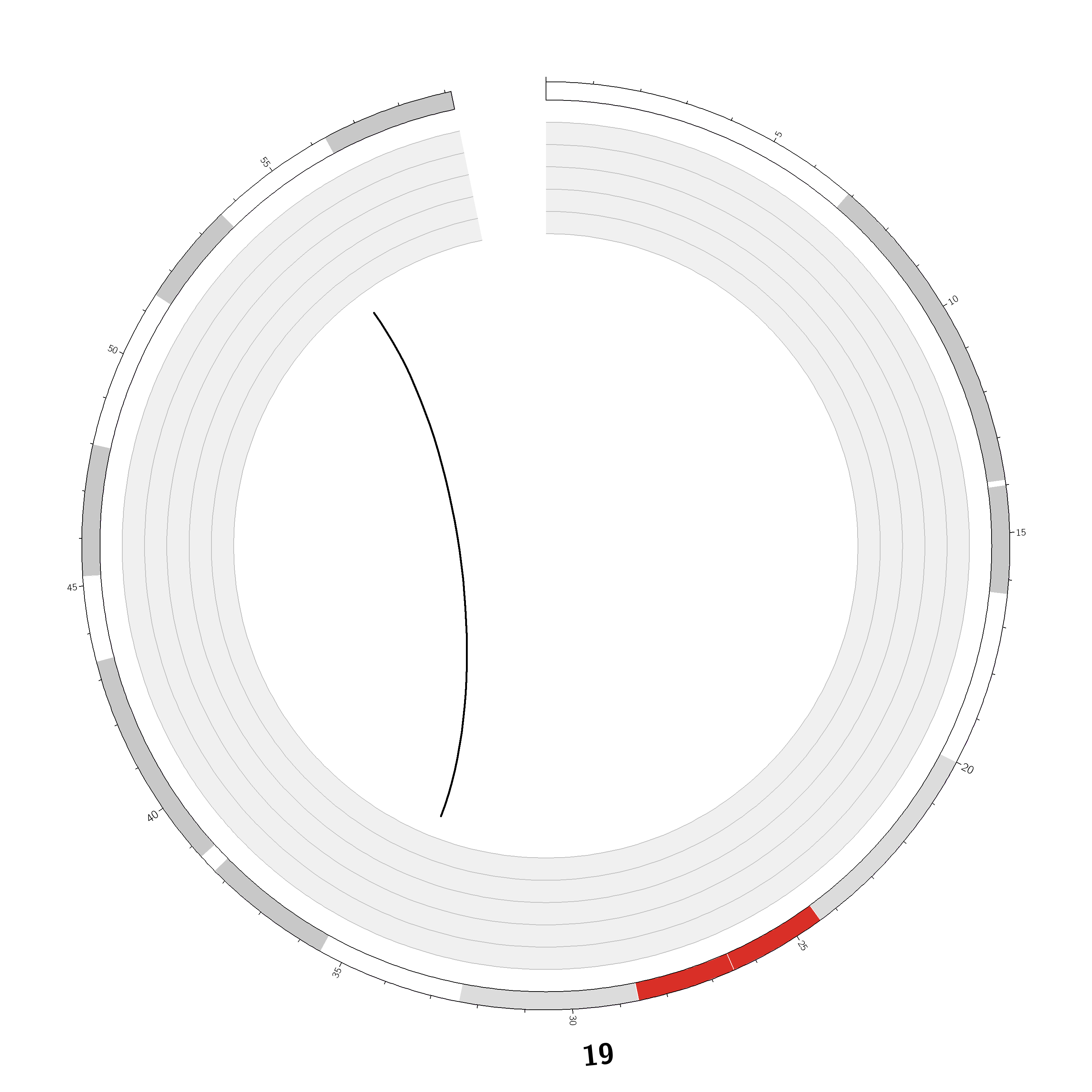
Case ID: |
TCGA-49-4488 |
| |
Chromosome: | 19 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| |
Affected Gene: |
Gene List |
| |
Download: |
Links File |
| |  | | TCGA-75-5146 |
| |
|
|
| |
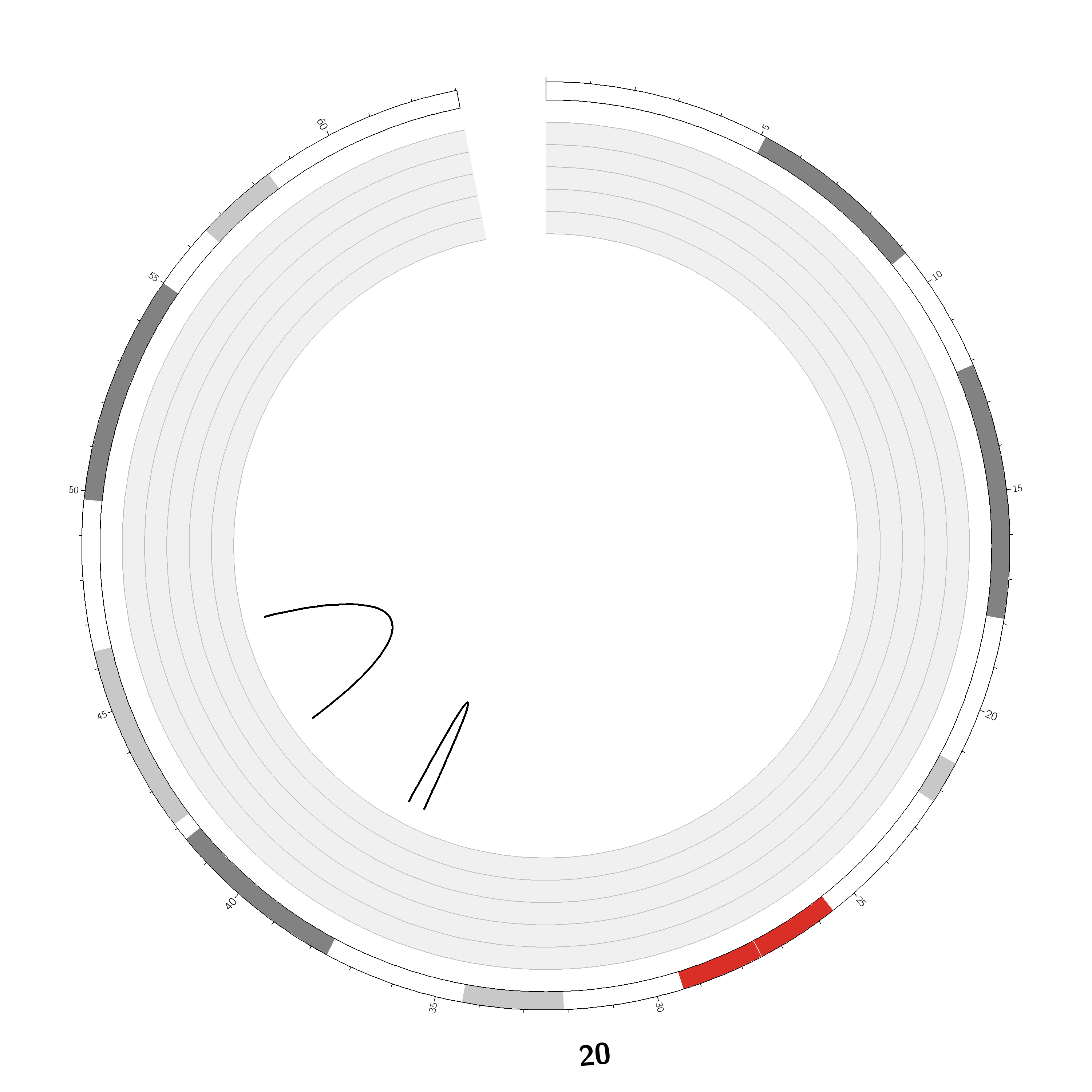
Case ID: |
TCGA-75-5146 |
| |
Chromosome: | 20 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| |
Affected Gene: |
Gene List |
| |
Download: |
Links File |
| |  | | TCGA-05-4422 |
| |
|
|
| |
Case ID: |
TCGA-05-4422 |
| |
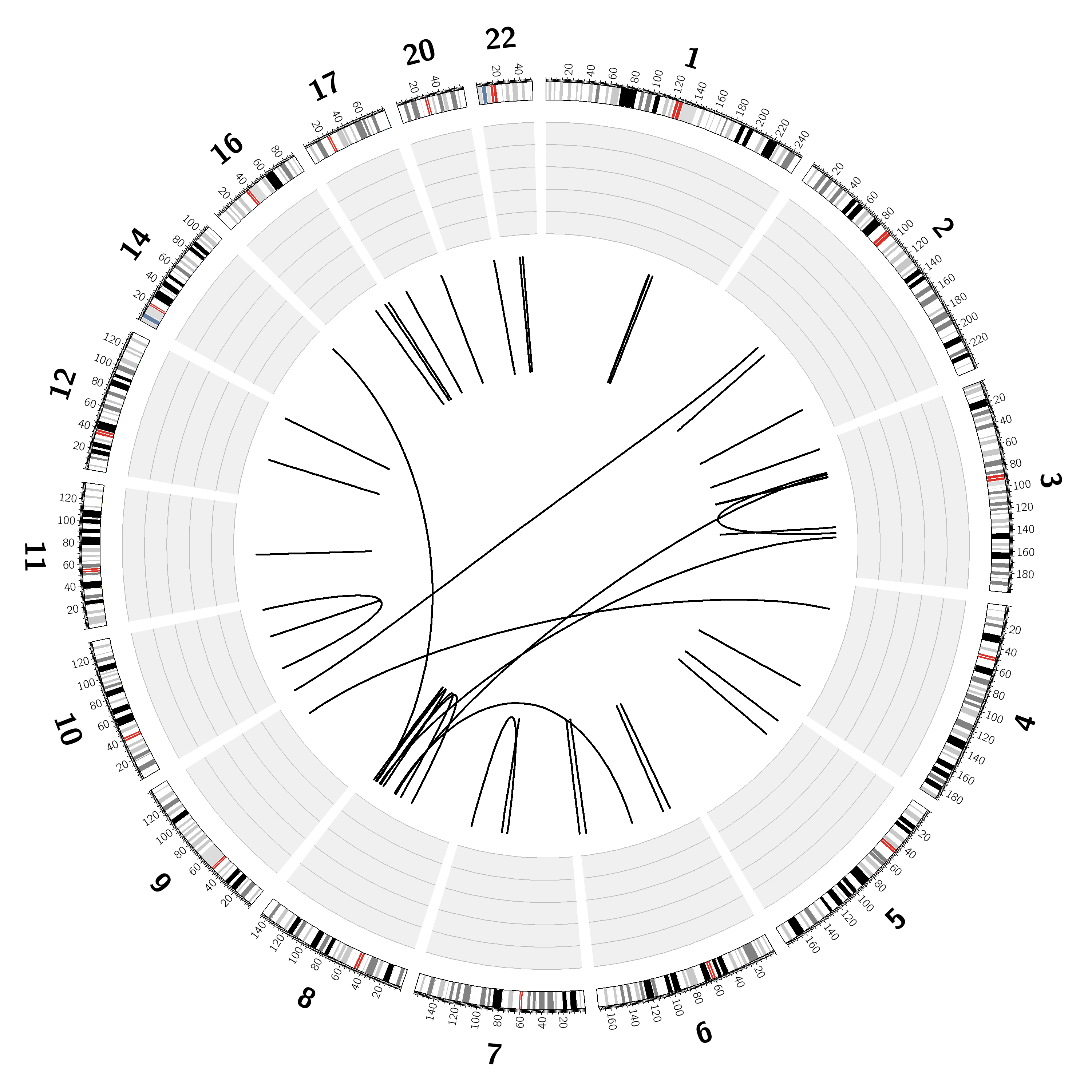
Chromosome: | 1,2,3,4,5,6,7,8,9,10,11,12,14,16,17,20,22 |
| |
Disease type: |
Non Small-cell lung cancer |
| |
Technology: |
Next Generation Sequencing |
| |
Platform: |
Illumina HiSeq2000 |
| |
Affected Gene: |
Gene List |
| |
Download: |
Links File |
| |  | | top |
|

